I have rendered three visual representations that depict the ATP synthase enzyme using Visual Molecular Dynamics software. ATP synthase, which can also be referred to as F-type ATPases, is an embedded-membrane ATP synthesizer enzyme that converts electrochemical energy into chemical energy; ATP, the standardized primary molecule for energy transfer and consumption at the cellular level. The enzyme can be found in the membrane of mitochondria (inner membrane), bacteria (plasma membrane), and chloroplasts (thylakoid membrane). With regards to its overall structure, ATP synthase is similar to transport ATPases, in particular the V-type pumps, however, ATP synthases are distinct from such ATP-driven pumps because ATP synthases use H+ through their membrane-embedded motor – F1 motor, please view images below – to drive the catalysis of the reaction creating the energy potential phosphate bond in ATP from ADP and phosphate.
The used atom coordinates for the ATP synthase, 5are, can be found in the references at the end of this writing, much appreciation to Zhou et al for deciphering this atomic material, and Humphrey et al for making VMD development available. The structural data used for the representation images created are ATP synthase molecules from Bovine mitochondria.
TODO: css responsive image resizing
Image 1 below, depicts the volumetric surface of the ATP synthase molecule, as well as, structural labels:
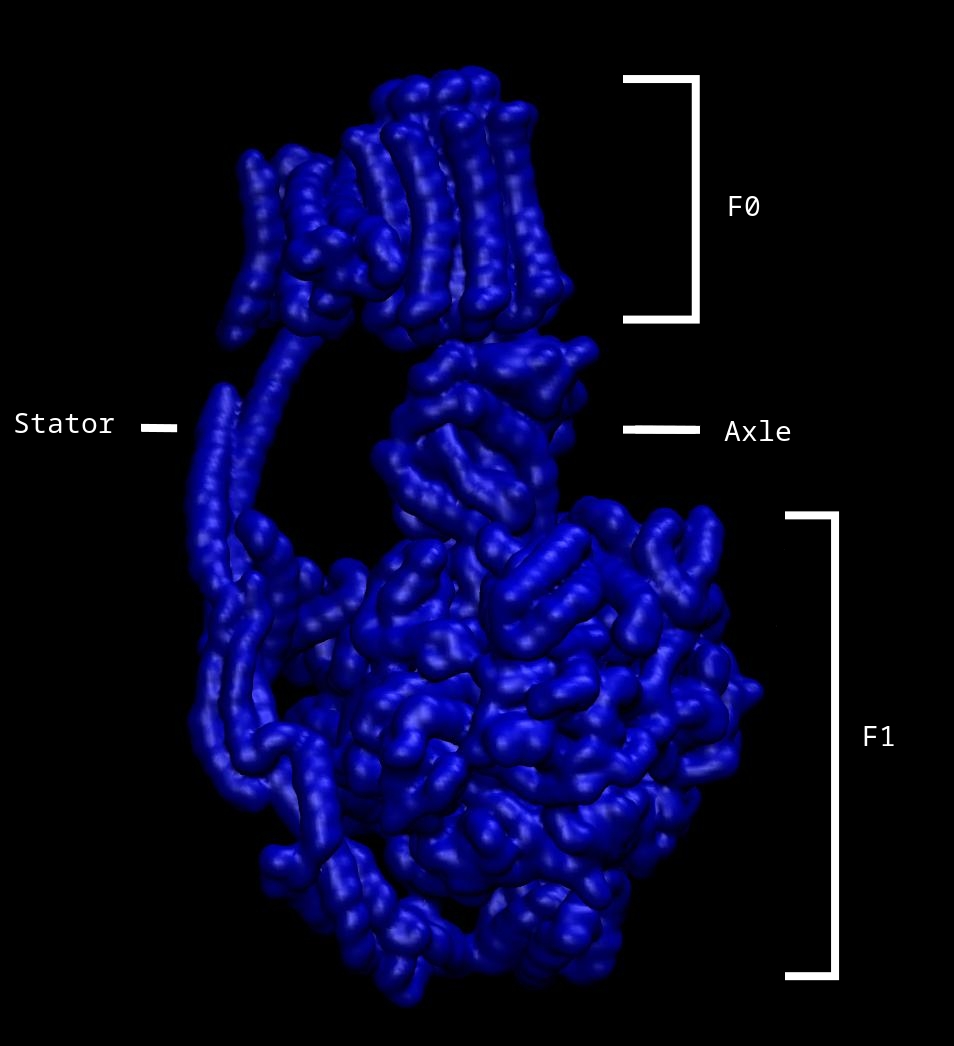
Image 2 below, depicts the same view of the ATP synthase molecule, however includes a specific internal representation detailing the structural outline of the complex within its volumetric possibilities – colouring denotes unique chains – where the axel is located, the grey chain, it roots down into the center of the F1 motor, so that its rotationary force can be shared:
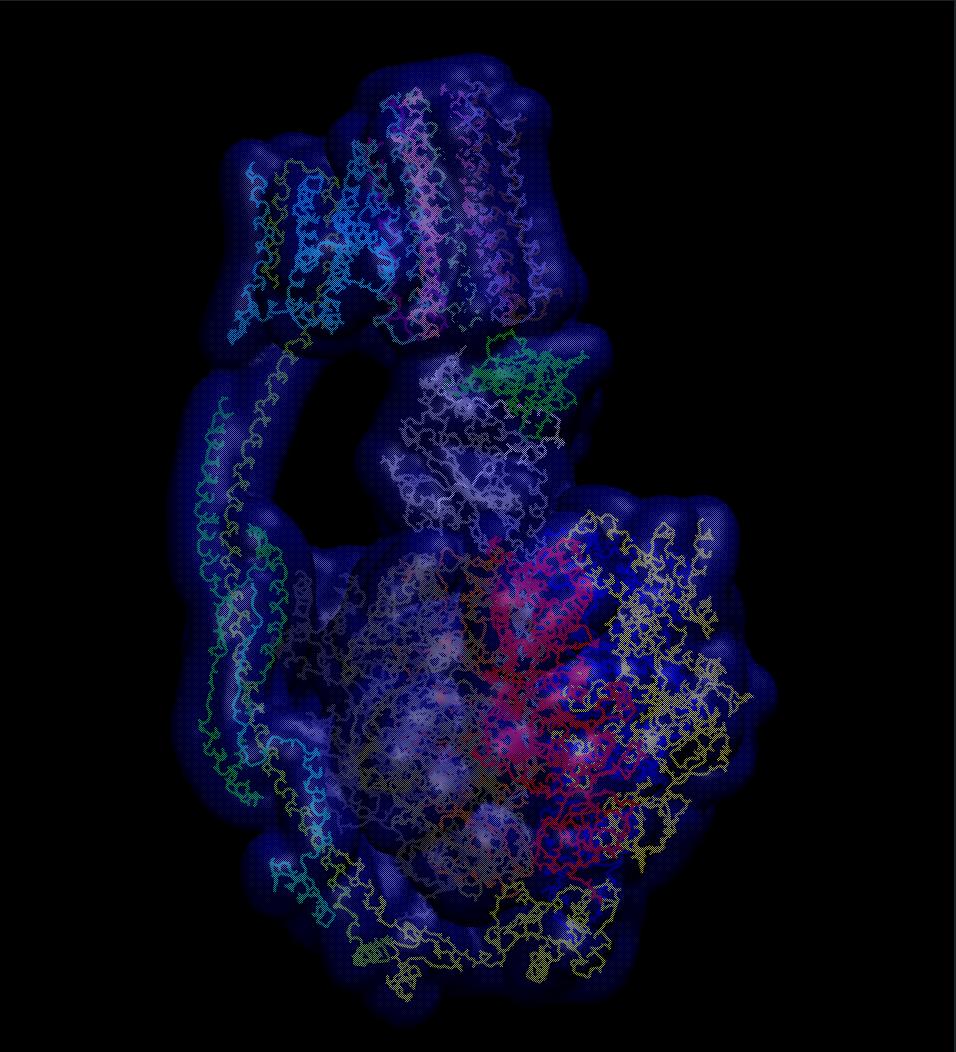
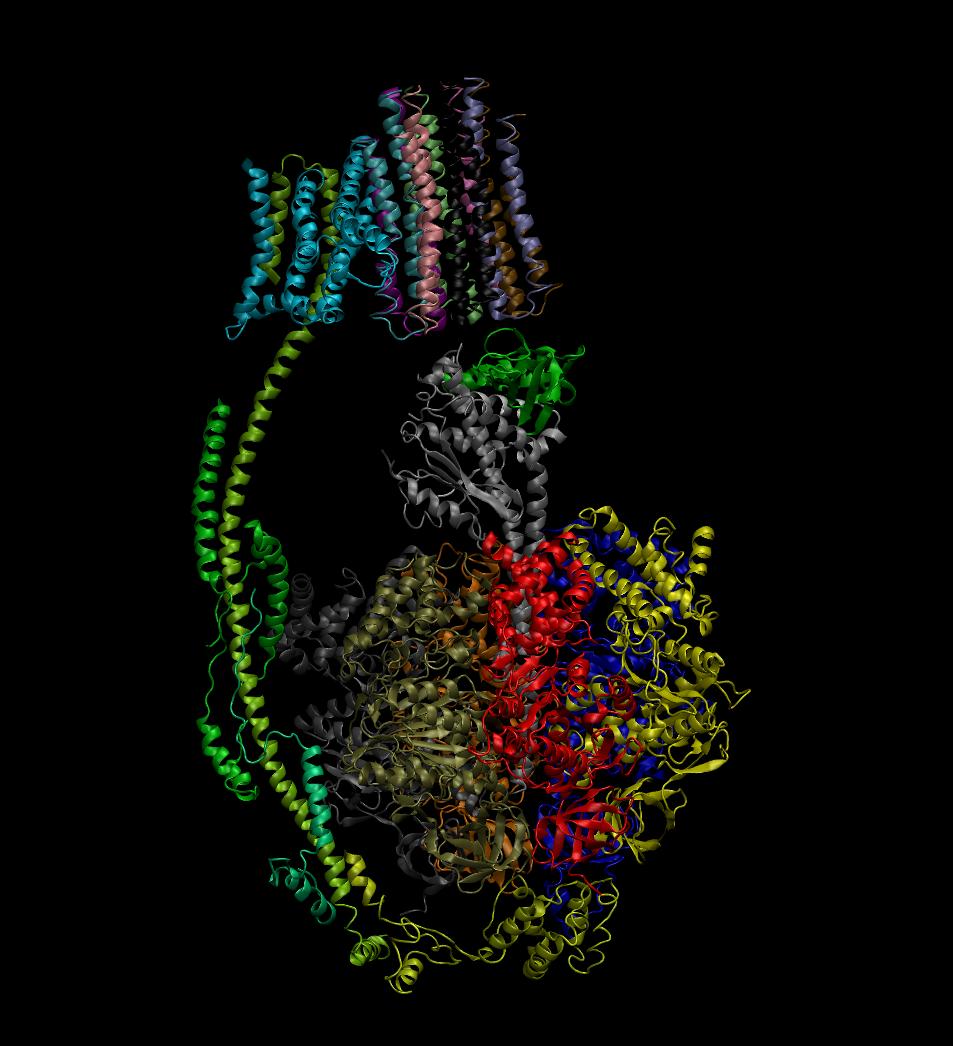
Image 3 below, depicts the same view of the ATP synthase molecule, the identity of the image draws the secondary features of the molecule where: spirals denote alpha-helix structures, thin tubular coils are turns, and flat wide arrows are beta-sheets. Structurally the molecule is composed of many more turns and alpha-helix structures than beta-sheets, and this likely due to the required flexibility the enzyme needs to complete rotations, whereas beta-sheets would be too rigid and restrictive for performing such tasks:
Labeled in Image 1, are the four main structural constituents of the membrane protein complex. Held together by the stator, the F0 motor is inserted within the membrane region and the F1 motor is found in the intracellular matrix, together forming the rotary motors of the ATP synthase molecule. The rotor subcomplex also consists of a asymmetric axle, and when H+ flows through the F0 motor, turning the cylindrical motor, the connected axle also turns. Additionally, as the axle is connected to the internal subunits of the F1 motor, turning of the axle causes structural changes to the F1 region which begins the catalysis of the reactionary process that forms the energy-rich, unstable phosphate to phosphate bond in ATP.
References
Humphrey, W., Dalke, A. and Schulten, K., “VMD - Visual Molecular Dynamics”, J. Molec. Graphics, 1996, vol. 14, pp. 33-38. Accessed: http://www.ks.uiuc.edu/Research/vmd/
5are: Zhou, A., Rohou, A., Schep, D. G., Bason, J. V., Montgomery, M. G., Walker, J. E., Grigorieff, N., & Rubinstein, J. L. (2015) Structure and conformational states of the bovine mitochondrial ATP syhnthase by cryo-EM. Elife, vol 4: e10180. DOI: 10.7554/eLife.10180 (PDB entry: https://www.rcsb.org/structure/5are)
Return Home

